Protein Domain Segmentor Tutorial
OS Compatibility: Mac OSX, Linux.
Prerequisites: PyMOL. Python 2.7.15 or later. Alternatively, Python 3.6.5 or later.
Download Link: http://github.com/egurapha/prot_domain_segmentor
BioArXiv: http://www.biorxiv.org/content/10.1101/474627v3
Datasets: All datasets from our manuscript are available HERE.
Step 1: Install Miniconda or Anaconda.
Please install the version of Miniconda or Anaconda needed for your version of Python: http://conda.io/en/latest/miniconda.html
Step 2: Download the Code Package from GitHub and run setup.sh
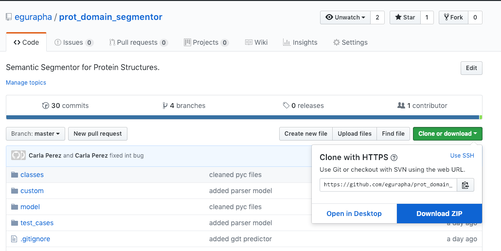
Download the software package from http://github.com/egurapha/prot_domain_segmentor by clicking the "Clone or Download" button.
Extract the Zip file in the desired location. The package requires the following
Python packages:
To install all of the necessary dependencies run the setup.sh file that comes with the code package by opening a terminal and typing "./setup.sh" in the prot_domain_segmentor folder. The package installation may take several minutes.
Prerequisites: PyMOL. Python 2.7.15 or later. Alternatively, Python 3.6.5 or later.
Download Link: http://github.com/egurapha/prot_domain_segmentor
BioArXiv: http://www.biorxiv.org/content/10.1101/474627v3
Datasets: All datasets from our manuscript are available HERE.
Step 1: Install Miniconda or Anaconda.
Please install the version of Miniconda or Anaconda needed for your version of Python: http://conda.io/en/latest/miniconda.html
Step 2: Download the Code Package from GitHub and run setup.sh
Download the software package from http://github.com/egurapha/prot_domain_segmentor by clicking the "Clone or Download" button.
Extract the Zip file in the desired location. The package requires the following
Python packages:
- Pytorch 0.4.1 or newer.
- Numpy 1.15.4 or newer.
- Scipy 1.1.0 or newer.
- BioPython 1.7.2 or newer
To install all of the necessary dependencies run the setup.sh file that comes with the code package by opening a terminal and typing "./setup.sh" in the prot_domain_segmentor folder. The package installation may take several minutes.

Step 3: Prepare an Input PDB.
Prepare a single chain PDB for input by opening a PDB in PyMOL and saving the chain as a new PDB file. Ligands and solvent present in the PDB will be ignored. Place the PDB in the prot_domain_segmentor folder. We have provided several example inputs in test_case folder .
Note for Large Proteins : The model can only return predictions for up to 512 residues at a time. For proteins greater than 512 residues, predictions for the center 512 residues in the amino acid sequence will be returned. For very large proteins we recommend making a best guess at where a domain boundary is, and then passing in fragments of the protein -- predictions for each fragment can be combined to obtain predictions for the full protein.
Step 4: Run the Model.
To run the model with PyMOL visualization, use the command:
pymol run_segmentor.py my_structure.pdb
The example output for the test case 3gqyA is shown on the right. The PyMOL terminal will report the corresponding architecture classes for each color.
The model can also be run without PyMOL, using the command:
python run_segmentor.py my_structure.pdb
This command will yield a text output of each the architecture classes found in the structure, and the residue numbers corresponding to each class.
The domain parser model can be run with the following commands in a similar fashion:
pymol run_parser.py my_structure.pdb
python run_parser.py my_structure.pdb
Updated: Raphael R. Eguchi. Feb 8, 2019
Prepare a single chain PDB for input by opening a PDB in PyMOL and saving the chain as a new PDB file. Ligands and solvent present in the PDB will be ignored. Place the PDB in the prot_domain_segmentor folder. We have provided several example inputs in test_case folder .
Note for Large Proteins : The model can only return predictions for up to 512 residues at a time. For proteins greater than 512 residues, predictions for the center 512 residues in the amino acid sequence will be returned. For very large proteins we recommend making a best guess at where a domain boundary is, and then passing in fragments of the protein -- predictions for each fragment can be combined to obtain predictions for the full protein.
Step 4: Run the Model.
To run the model with PyMOL visualization, use the command:
pymol run_segmentor.py my_structure.pdb
The example output for the test case 3gqyA is shown on the right. The PyMOL terminal will report the corresponding architecture classes for each color.
The model can also be run without PyMOL, using the command:
python run_segmentor.py my_structure.pdb
This command will yield a text output of each the architecture classes found in the structure, and the residue numbers corresponding to each class.
The domain parser model can be run with the following commands in a similar fashion:
pymol run_parser.py my_structure.pdb
python run_parser.py my_structure.pdb
Updated: Raphael R. Eguchi. Feb 8, 2019